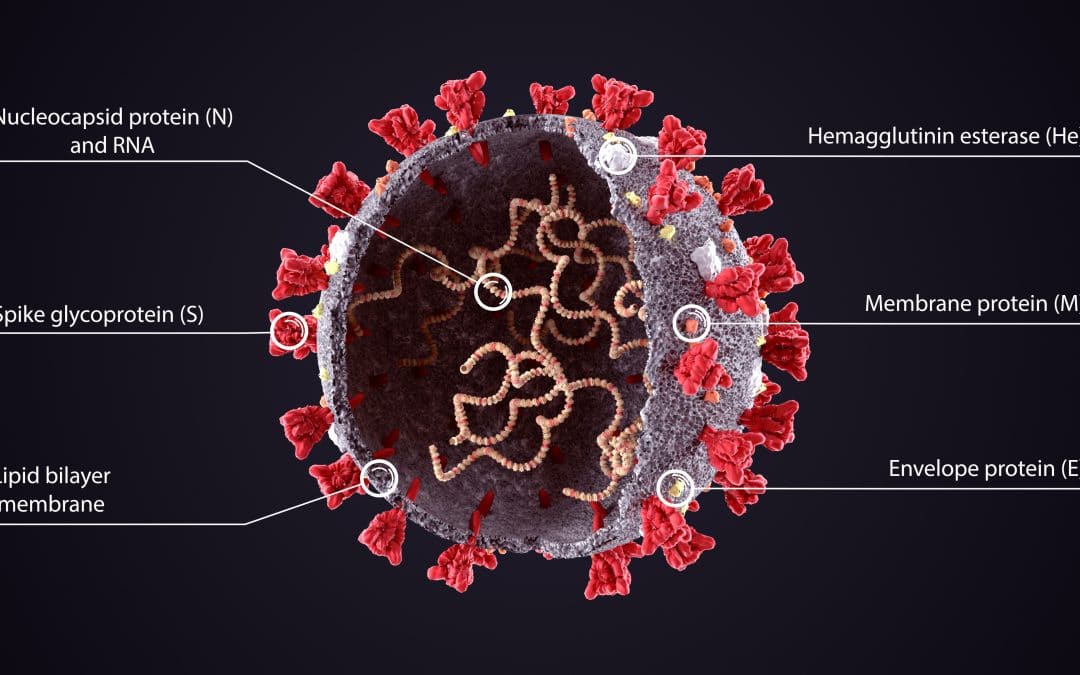
Another new COVID-19 variant of concern has emerged. A variant is labelled “of concern” when it spreads more easily and is more virulent or resistant to vaccines. How does infrastructure help scientists detect new COVID-19 variants? As a technologist, do you wonder what architecture supports these very important applications?
Omicron emerges
The Africa Centres for Disease Control and Prevention raised the alarm about a new variant. This variant is now named Omicron. Fortunately the African team was able to quickly identify a new variant. More importantly, they immediately shared their scientific observations with the world community of virologists, hoping the new data could be used to prevent another deadly wave of infection.
Here are some of the African agency’s observations:
- By 11/25, the new variant had been detected in 77 samples. These samples were collected between 11/12 to 11/20 from Gauteng province in South Africa, Botswana and Hong Kong.
- The variant displays multiple mutations across the virus genome, including more than 30 in the region which encodes the spike protein responsible for virus entry into host cells.
- Many of the identified mutations are not yet well characterized and have not been identified in other currently circulating variants.
Don’t you wonder how they are able to figure this out so quickly?
How does the COVID-19 virus work?
Time for a biology lesson! Viruses hack the machinery of cells, per virologist Rich Condit. I love this presentation. Dr Condit describes biology as we would describe infrastructure.
How does technology help scientists find new mutations?
I found it helped to listen to Dr. Condit’s virology lesson. It helped me wrap my mind around how an application used to find viral mutations may need to be architected. For instance, Dr. Condit explains the genetic makeup of viruses. In fact, he provides detail about the genetic makeup of the SARs-CoV-2 virus (~ 14:00). Understanding that each strain of the virus has its own genetic markers makes it obvious which technical tools could be used to find new variants.
For example, here’s one scenario of how SARs-CoV-2 variants are found. If you take a COVID-19 test and it’s positive for the virus, your sample may be screened with a genotyping panel. That screening panel may include a SNP (Single Nucleotide Polymorphisms) identification pipeline.
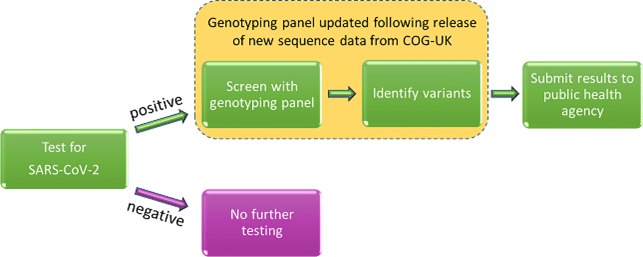
Detecting SARS-CoV-2 variants with SNP genotyping. via “Detecting SARS-CoV-2 variants with SNP Genotyping”.
SNP genotyping identifies genetic variations using sequenced SARs-CoV-2 samples. For example, the genotyping test can detect if the version of the virus that infected you is genetically different than known versions of the virus. Next, if a fast growing cluster of the new variant is found, health officials can be alerted. Finally, scientists will work to understand how the new genetic sequence of the virus works (how virulent it is, if there will be vaccine break-thru, etc.).
The scientists get to work
Currently, scientists are learning more about the Omicron variant. South African scientists detected the variant. When they noticed a cluster of infections, they informed WHO (the World Health Organization). WHO advised countries to enhance surveillance and genomic sequencing efforts. Additionally, scientists were asked to share any new information they find with GISAID.
GISAID is a global database platform for virologists. It was built to enable rapid and open access to epidemic and pandemic virus data. Scientists use it to share data. First, they upload protein or nucleotide sequences in a FASTA file. Next, the server guides them to the correct sequences to compare.
I can’t lie, the language is extremely scientific. However, if you’re a scientist who understands the genetic structure of a virus, you’ll probably understand the instructions. Incredibly this data is available in near real time. Of course, in a global pandemic reducing the time it takes to find solutions is a literal life or death matter.
CLIMB-BIG-DATA project
Now that we have an idea of how applications used to find new SARs-CoV-2 variants may work we can think about required infrastructure. One example is the CLIMB-BIG-DATA project (the Cloud Infrastructure for Big Data Microbial Bioinformatics). CLIMB is a collaboration of UK universities. The project helps scientists solve big data projects for bioinformatics. It most likely is the largest single system dedicated to Microbial Bioinformatics research in the world.
This page lays out the system highlights:
- It runs on OpenStack (on IBM supplied hardware)
- Over 7500 CPU cores
- 78TB RAM
- Ceph object storage
- 2304 TB cross site replicated storage
- 27 Dell R730XD servers per site, each with 16x 4TB HDDs
- 500 TB local storage (IBM GPFS)
- Brocade networking
- 1000 VMs
- Users log into access VMs that are pre-configured for microbial genome analysis
- Root access available so users can install their own software
- User VMs can access resources such as Genomics Virtual Laboratory and Galaxy
Real Talk
It is easy to forget why we build infrastructure. While we nerd out on how to manage and move data around, our infrastructures support science that is equally difficult to understand. I’m sure there are many other examples of architectures that are supporting work being done to contain this pandemic.
Spend some time nerding-out with your users. Maybe what you learn will lead to the next infrastructure innovation!
Featured image via Shutterstock.